Professor Rozners has delivered invited lectures at various national and international conferences, major research universities as well as small colleges. If you are interested in hosting a lecture at your school please contact Professor Rozners directly.
Lecture Abstracts
Amide-Modified Oligonucleotides for Chemical Control of Functional RNAs
RNA-based technologies to control gene expression, such as, RNA interference (RNAi) and CRISPR have become powerful tools in molecular biology and genomics. The exciting potential that RNAi and CRISPR may also become new therapeutic approaches has reinvigorated interest in chemically modifying RNA to improve its properties for in vivo applications. Chemical modifications can improve enzymatic stability, in vivo delivery, cellular uptake, and sequence specificity; as well as minimize off-target activity of short interfering RNAs (siRNAs) and CRISPR associated RNAs (crRNAs). The long-term goal of our research is to develop chemical modifications for optimization of in vivo potential of siRNAs and crRNAs. Our current work is focused on the development of nonionic analogues of RNA that have the phosphodiesters replaced by amide linkages (AM1 in Figure) [1].
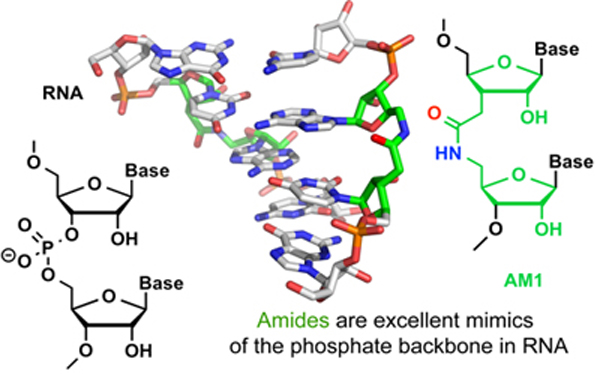 We hypothesize that the reduced negative charge and conformational preferences of amide linkages can optimize potency, cellular uptake, and reduce off-target effects of siRNAs and crRNAs. This presentation will discuss synthesis, structure, and RNAi and CRISPR activity of amide-modified RNA. Structural studies show that amides are excellent mimics of the phosphodiester inter-nucleoside linkages in RNA [2]. The local conformational changes caused by the amide linkages where accommodated easily by small adjustments in RNA structure. RNAi activity assays show that amides are well tolerated at internal positions in both strands of siRNAs. Surprisingly, amide modifications in the middle of the guide strand and at the 5´-end of the passenger strand increased the RNAi activity compared to unmodified siRNA [1]. Most remarkably, replacement of certain phosphate linkages with amides significantly reduced the off-target activity of guide and passenger strands [3]. Most recent studies by our group showed that amide modifications did not interfere with CRISPR-Cas9 activity when placed in the protospacer adjacent motif distal region of crRNAs [4]. In contrast, modification of the seed region of crRNAs led to loss of DNA cleavage activity at most but not all positions. Taken together, our results suggest that amides are excellent mimics of phosphate backbone in RNA and may have potential to optimize biological and pharmacological properties of siRNAs and crRNAs for in vivo applications.
We hypothesize that the reduced negative charge and conformational preferences of amide linkages can optimize potency, cellular uptake, and reduce off-target effects of siRNAs and crRNAs. This presentation will discuss synthesis, structure, and RNAi and CRISPR activity of amide-modified RNA. Structural studies show that amides are excellent mimics of the phosphodiester inter-nucleoside linkages in RNA [2]. The local conformational changes caused by the amide linkages where accommodated easily by small adjustments in RNA structure. RNAi activity assays show that amides are well tolerated at internal positions in both strands of siRNAs. Surprisingly, amide modifications in the middle of the guide strand and at the 5´-end of the passenger strand increased the RNAi activity compared to unmodified siRNA [1]. Most remarkably, replacement of certain phosphate linkages with amides significantly reduced the off-target activity of guide and passenger strands [3]. Most recent studies by our group showed that amide modifications did not interfere with CRISPR-Cas9 activity when placed in the protospacer adjacent motif distal region of crRNAs [4]. In contrast, modification of the seed region of crRNAs led to loss of DNA cleavage activity at most but not all positions. Taken together, our results suggest that amides are excellent mimics of phosphate backbone in RNA and may have potential to optimize biological and pharmacological properties of siRNAs and crRNAs for in vivo applications.
References
- Kotikam, V.; Rozners, E. Amide-Modified RNA: Using Protein Backbone to Modulate Function of Short Interfering RNAs Acc. Chem. Res. 2022, 53, 1782-1790.
- Selvam, C.; Thomas, S.; Abbott, J.; Kennedy, S. D.; Rozners, E. Amides Are Excellent Mimics of Phosphate Linkages in RNA Angew. Chem. Int. Ed. 2011, 50, 2068-2070.
- Richter, M.; Viel, J. A.; Kotikam, V.; Gajula, P. K.; Coyle, L.; Pal, C.; Rozners, E. Amide Modifications in the Seed Region of the Guide Strand Improve the On-Target Specificity of Short Interfering RNA ACS Chem. Biol. 2023, 18, 7-11.
- Kotikam, V.; Gajula, P. K.; Coyle, L.; Rozners, E. Amide Internucleoside Linkages Are Well Tolerated in Protospacer Adjacent Motif-Distal Region of CRISPR RNAs ACS Chem. Biol. 2022, 17, 509-512.
Sequence Selective Recognition of Double-Stranded RNA Using Modified Peptide Nucleic Acids
The important roles that non-coding double-stranded RNAs (dsRNA) play in biology and development of disease makes them attractive targets for molecular recognition. However, designing ligands that sequence specifically recognize complex folded RNA has been a challenging and involved process because RNA helix presents little opportunity for shape-selective recognition. We discovered that cationic nucleobase- and backbone-modified peptide nucleic acids (PNA) bind with high sequence selectivity and low nanomolar affinity to dsRNA via major groove triple helix formation under physiological conditions [1]. Most interestingly, the modified PNAs exhibited unique RNA selectivity and had up to two orders of magnitude higher affinity for the dsRNAs than for the same dsDNA sequences. NMR structural studies showed that PNA and RNA formed a hydrogen bonding zipper that stabilized the triple helix, which was not possible in PNA-dsDNA complex due to longer inter-phosphate distances of DNA.
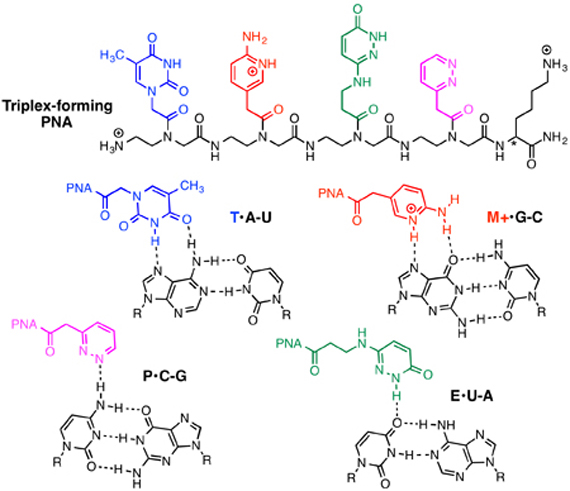 Nucleobase- and backbone-modified PNAs recognized dsRNA sequences present in biologically relevant RNA, such as microRNAs, with high affinity under physiological conditions. In collaboration with Prof. Naoki Sugimoto’s group at FIBER, Kobe, Japan, we demonstrated that binding of triplex-forming PNA inhibited maturation of pre-microRNA hairpins [2]. Recently, we discovered that triplex-forming PNAs had unusually high affinity for single purine nucleotide bulges in double-helical RNA. Binding of triplex-forming PNA switched the conformation of single purine bulges in dsRNA between stacked-in and looped-out depending on the sequence of PNA [3].
Taken together our results suggest that the cationic modified PNAs may be promising compounds for modulating the function of biologically relevant dsRNA in live cells. The ability to control dynamic equilibria of RNA’s structure will be an important tool for studying structure-function relationships in RNA biology and may have potential in novel therapeutic approaches targeting disease related RNAs. The current presentation will discuss our most recent results on sequence selective recognition of complex biological dsRNA molecules and potential applications of this recognition in biomedical research and biotechnology [4].
Nucleobase- and backbone-modified PNAs recognized dsRNA sequences present in biologically relevant RNA, such as microRNAs, with high affinity under physiological conditions. In collaboration with Prof. Naoki Sugimoto’s group at FIBER, Kobe, Japan, we demonstrated that binding of triplex-forming PNA inhibited maturation of pre-microRNA hairpins [2]. Recently, we discovered that triplex-forming PNAs had unusually high affinity for single purine nucleotide bulges in double-helical RNA. Binding of triplex-forming PNA switched the conformation of single purine bulges in dsRNA between stacked-in and looped-out depending on the sequence of PNA [3].
Taken together our results suggest that the cationic modified PNAs may be promising compounds for modulating the function of biologically relevant dsRNA in live cells. The ability to control dynamic equilibria of RNA’s structure will be an important tool for studying structure-function relationships in RNA biology and may have potential in novel therapeutic approaches targeting disease related RNAs. The current presentation will discuss our most recent results on sequence selective recognition of complex biological dsRNA molecules and potential applications of this recognition in biomedical research and biotechnology [4].
References
- Zengeya, T.; Gupta, P.; Rozners, E. Triple Helical Recognition of RNA Using 2-Aminopyridine-Modified PNA at Physiologically Relevant Conditions. Angew. Chem., Int. Ed. 2012, 51, 12593-12596.
- Endoh, T.; Brodyagin, N.; Hnedzko, D.; Sugimoto, N.; Rozners, E. Triple-Helical Binding of Peptide Nucleic Acid Inhibits Maturation of Endogenous MicroRNA-197. ACS Chem. Biol. 2021, 16, 1147-1151.
- Ryan, C. A.; Rahman, Md M.; Kumar, V.; Rozners, E. Triplex-Forming Peptide Nucleic Acid Controls Dynamic Conformations of RNA Bulges. J. Am. Chem. Soc. 2023, 145, 10497–10504.
- Katkevics, M.; MacKay, J. A.; Rozners, E. Triplex-forming peptide nucleic acids as emerging ligands to modulate structure and function of complex RNAs. Chem. Commun. 2024, 60, 1999-2008.
